Whole genome sequencing is a technique which determines the entire sequence of the whole genome of an organism. The genome is the complete genetic information of an organism. Here is what you need to know about the history and new advances of the whole genome sequencing technology.
Background information
As previously explained, the genome is the complete genetic information of an organism. Most organisms have DNA genomes, although there are known viruses with RNA genomes too. The genome of Eukaryotes (organisms with complex cells, i.e. cats and humans) consists of the cellular DNA (in the nucleus of the cell), and DNA placed in the organelles (mitochondria and chloroplasts).
Whole genome sequencing is the method of determination of the entire sequence of the whole genome of an organism. The DNA is made from two chains of nucleotides. Every nucleotide carries one of four possible bases (adenine, guanine, thymine or cytosine). The information within the genomic sequence is deciphered as the order of these four bases in the DNA chain (Figure 1). When we know a DNA sequence, we can analyze it and compare it to other sequences of the same or other organism.

Figure 1: DNA model sequence. Nucleotides carrying the four bases are color-coded (cytosine – blue, guanine – green, adenine – yellow and thymine – red) (Figure credits).
History
The very first method for genome sequencing was Maxam-Gilberts sequencing method developed in 1976 by Allan Maxam and Walter Gilbert. In 1977, Frederick Sanger and colleagues developed the Sanger sequencing method which was the most widely used sequencing method for about 40 years. These methods were manual, slow and allowed the sequencing of short DNA segments only. Today automated, more rapid techniques have been developed that allow sequencing of longer DNA strands.
The first complete genome to ever be sequenced was the genome of Haemophilus influenzae (a Gram-negative, pathogenic bacteria) in 1995. Shortly, many other microorganisms followed suit. The first eukaryotic genome to be sequenced was that of Saccharomyces cerevisiae (the yeast) in 1996. The first sequenced multicellular eukaryote, however, was the nematode worm Caenorhabditis elegans in 1998.
Human genome was sequenced as a part of The Human Genome project. Started in 1990, The Human Genome project was declared complete in 2003. That is, as complete as it can be, given the available technology. “The human genome has not been completely sequenced and neither has any other mammalian genome as far as I’m aware,” said Harvard Medical School bioengineer George Church.
First canine genome was sequenced and published in 2004, followed up with feline genome in 2007.

Techniques used for whole genome sequencing
Due to the large size of mammalian genomes, long DNA strands are first broken down into smaller portions, sequenced and then put together using bioinformatics methods. One of the most commonly used methods for this is whole-genome shotgun method, which was also partially used for The Human Genome project. New modern techniques such as capillary sequencing, illumina dye sequencing, pyrosequencing, SMRT sequencing and nanopore technology are all emerging and developing as well.
This method is used specifically for long strands of DNA. The DNA is randomly cut into numerous shorter fragments for sequencing (usually 2-150 kb long). These short fragments are then cloned into vectors (a vector is a DNA molecule used as a “vehicle” to artificially carry foreign genetic material into another cell, where it can be replicated). The cloned short fragments are now sequenced starting from both ends using the chain termination method. Each sequence is called “read.” The original sequence is afterward reconstructed from the reads using sequence assembly software.
In hierarchical shotgun, prior to actual sequencing, a physical map of the genome is made. This allows the planning of a minimal number of cut fragments that will cover the entire chromosome. Reducing the number of fragments reduces the amount of sequencing and assembly required for the process to be carried out.
To assembly the fragments, it is first determined in which way the clones overlap. After the identification of the overlaps, the sequences can be combined to construct the genome consensus (Figure 2).


Figure 2: In whole genome shotgun sequencing (a), the entire genome is randomly cut into small fragments and then reassembled. In hierarchical shotgun sequencing (b), the genome is first broken into larger segments. After the order of these segments is deduced, they are further cut into small fragments (Figure credits).
Sequencing the genome makes it possible for us to identify mutations and estimate mutation frequencies too. Thousands of pathogenic mutations have been identified through GS in recent years. Identification of mutations is very important because it reveals the genetics behind many genetically determined conditions and disorders. Estimation of mutation frequencies can also be very informative.
For example, it is known that the mutation rate in cancer is significantly higher than in healthy tissues due to genome instability. We also learned that the mutation rate isn’t the same across the regions of genome. Gene-rich regions undergo fewer mutations than non-coding regions, possibly due to DNA repair activity that is higher and more precise in these regions.
Information deciphered by WGS, such as the example above, allows unique insights into the roles and nature of different genes and genetic regions. This can subsequently be utilized in many different applied sciences.

Feline genome
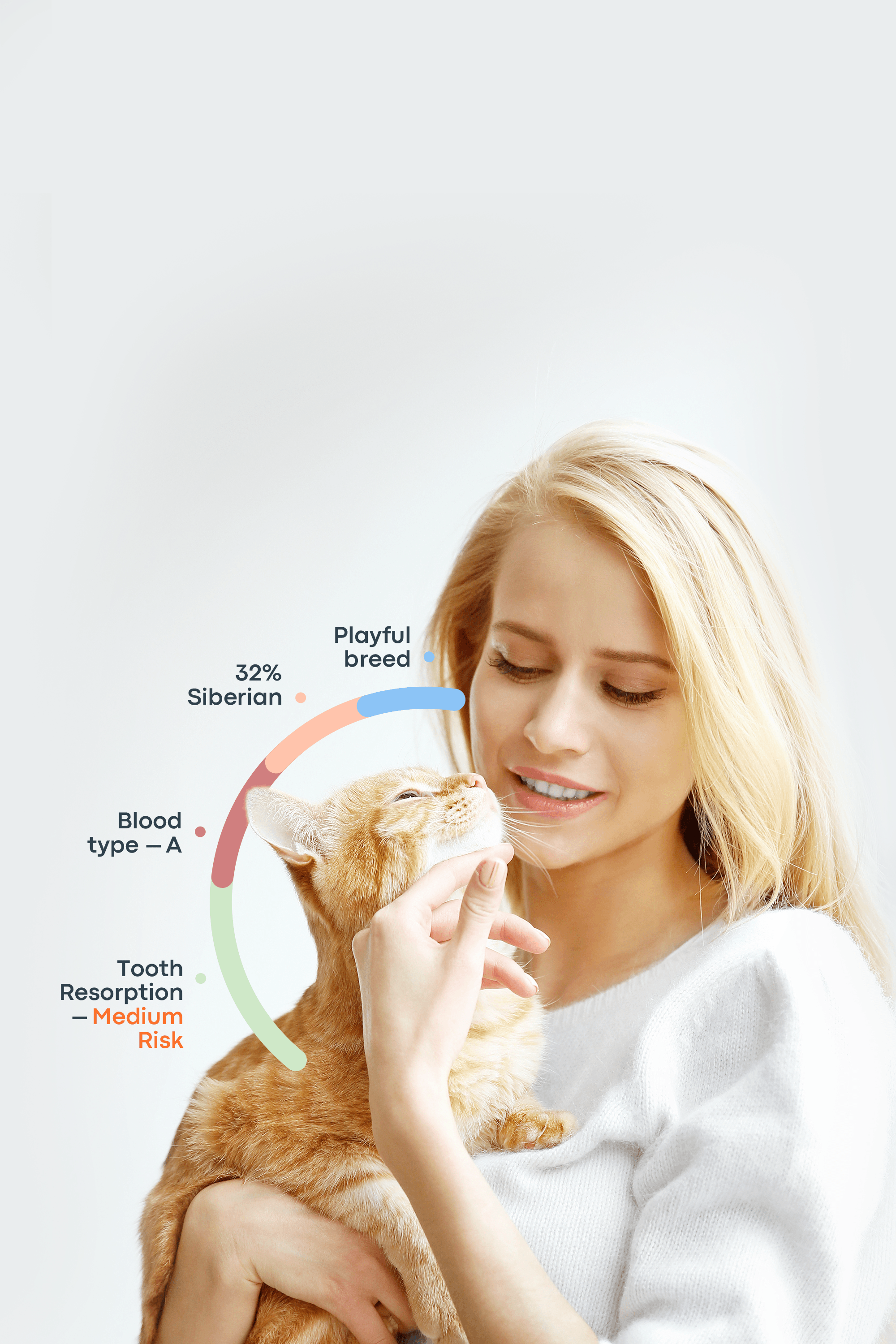
Cats have 38 chromosomes (for comparison, humans have 46), and roughly 20,000 genes. However, in order to make new discoveries and conclusions about cat DNA, the complete feline genome had to be sequenced first. Feline genome was finally sequenced for the first time in 2007! The cat whose genome was sequenced was a 4-year-old Abyssinian named Cinnamon. This allowed countless new discoveries in the field of feline genomics.
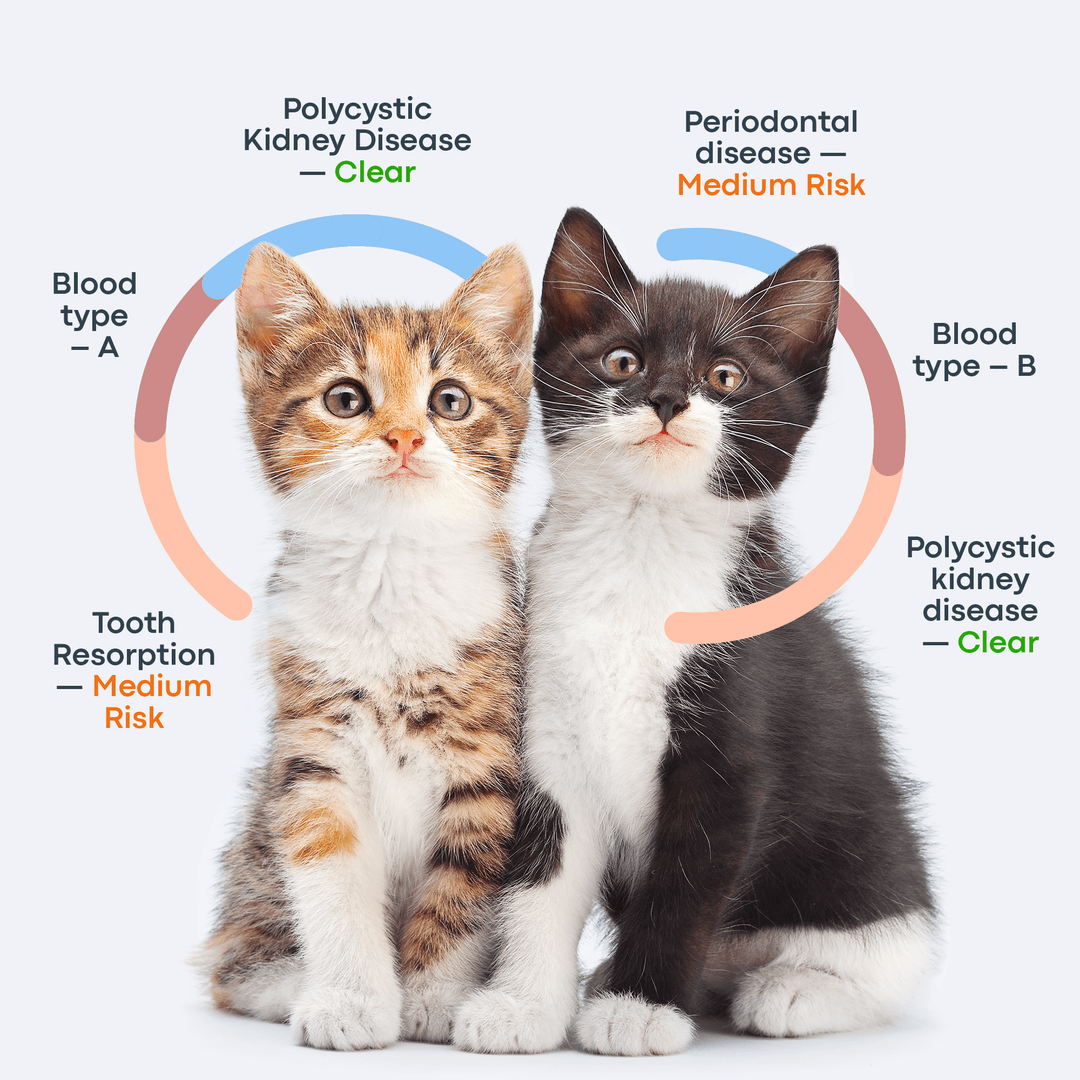
So far, about 250 genetic disorders have been found in cats. With the mapped sequences, these diseases can be further studied in order to be fully described and assessed.
Another intriguing discovery upon feline genome description was that humans share about 90% of DNA with felines! This makes cats genetically closer to humans than dogs. The genome sequence analysis also showed that humans and cat lineages diverged about 100 million years ago.
These and many other pieces of information enable us to learn more about cats and provide them with a better health system and support. Our felines can also serve as excellent models for human diseases due to high genetic resemblance. There are actually over 200 hereditary diseases which closely correlate between humans and cats. Among the most serious diseases common to cats and humans are leukemia, Alzheimer’s, HCM and HIV. Advances in research of these diseases in cats coulddirectly lead to advances in human medicine too.
Here at Basepaws, WGS enables us to do breed ancestry analysis, wildcat ancestry analysis and genetic screening for health markers.

Whole genome sequencing enables us to understand life at the molecular level. There is so much more WGS can still teach us about our own genome, as well as the feline. We can’t even begin to imagine the power of this knowledge. The potential is superior.
Improvement of this technology will open countless doors to many scientific fields such as personalized medicine, nutrition, pathology, microbiology, evolution, pharmacy, agrigenomics and numerous other fields. We are only yet starting to realize the full potential of WGS. How exciting!